User Tools
Sidebar
Table of Contents
An introduction to Jas4pp is given in this Snowmass21 contribution: S.V. Chekanov, G. Gavalian and N. A. Graf, “Jas4pp - a Data-Analysis Framework for Physics and Detector Studies” arXiv:2011.05329, Comp. Physics. Comm. 262 (2021) 107857, ANL-HEP-164101, SLAC-PUB-17569
Show the BibTex entry for this article here
Show the BibTex entry for this article here
@article{Chekanov:2020bja,
author = "Chekanov, S. V. and Gavalian, G. and Graf, N. A.",
title = "{Jas4pp -- a Data-Analysis Framework for Physics and Detector Studies}",
eprint = "2011.05329",
archivePrefix = "arXiv",
primaryClass = "physics.comp-ph",
reportNumber = "ANL-HEP-164101, SLAC-PUB-17569",
doi = "10.1016/j.cpc.2021.107857",
journal = "Comput. Phys. Commun.",
volume = "262",
pages = "107857",
year = "2021"
}
Jas4pp manual
Jas4PP program is a data-analysis environment for detector and physics studies of future circular colliders. Jas4PP is a merge of several open-source Java projects, such as
- lcsim.org developed at SLAC
- Many file formats used in HEP, including ROOT5 / 6(partially). See Section supported file formats.
- GROOT and XROOTD - data visualization and I/O used at JLab
- j4np-physics - Physics vectors from JLab
- FreeHep developed at SLAC
- DataMelt (community edition) from jWork.ORG
- Additional libraries such as Apache Common Nath3, JFreeChart and other.
- Various physics packages for statistics, limits settings etc.
The program is designed to work using the following languages:
- Java (+8 version)
- Python language using Jython 2.7.2 (which is compatible with the 2.7.2 version of CPython)
- Groovy (version 3.0.6) https://groovy-lang.org/
Installation
The installation does not have external dependencies. To run Jas4pp, you need to install Java. To check you Java installation, run “java -version”. One can find Oracle JDK or JRE versions from the Oracle download link. OpenJDK can be downloaded from https://openjdk.java.net/.
You can download the Jas4pp program using the download page.
You can use Jas4pp installer program (jas4pp-[VERSION]-installer.jar). It asks several questions and creates an icon on a desktop. It is recommended to install the program in “C:\Jas4pp” (for Wnindows) to avoid permission problems.
Alternatively, one can use untar the program in any directory. Run these commands to install the package using Linux/Mac with the “bash” shell:
bash> wget https://atlaswww.hep.anl.gov/asc/jas4pp/download/current.php -O jas4pp.tgz bash> tar -zvxf jas4pp.tgz bash> cd jas4pp bash> source ./setup.sh # takes 5 sec for first-time optimization bash> jaspp
The last command starts Jas4PP GUI:
How to run
Use this GUI to explore examples with data visualisation shown in the main window: [JAS4PP examples]-[Python examples] and then [F2] (or use the mouse menu) to execute:
If you run Jython/Groovy code in a batch mode, use the following commands:
bash> fpad code.py # execution of Python/Jython code bash> fpad code.groovy # execution of Groovy code
You are ready to run over any file with truth-level (ProMC or LCIO files) and detector-simulation information (LCIO files) from HepSim repository. Here is a list of Jas4pp commands:
jaspp # start Jas4pp with preconfigured HEP plugins (Linux/Mac with bash) jaspp.bat # same for Windows fpad # run Jython scripts in a batch mode (Linux/Mac with bash) fpad_edit # start a minimalistic Python editor "FPAD") and Jython shell fpad_edit.bat # same for Windows gconverter # geometry converters gconverter_gui # same in GUI mode hs-help # shows commands to access data from the HepSim repository
Use these programs for:
- Downloading and searching HepSim data
- Processing ProMC files from HepSim
- Running over SLCIO files with Geant4 simulated / reconstructed events.
- Data analysis (jets, physics vectors, histogram packages, limit settings, etc.)
- Visualization of reconstructed events using Wired4 display
You can find more details in HepSim web page.
We recommend to use Groovy for analysis programs. One advantage of Groovy is that the code execution for large loops is a factor 12 faster than for Jython and CPython). See the benchmark. All HEP Jas plugins are included (and some of them were modified to work with DataMelt).
Jas4pp can be extended by putting external jar files in the directory “lib/user”, or linking pure-Python modules.
Examples
All examples of Jas4pp are collected in the directory “examples”. Run them as:
fpad examples/dmelt/histo1.py # DMelt histogram example fpad examples/groot/histogram1d.py # GROOT histogram example fpad examples/jaida/Fit.py # Jas/Aida fit example fpad examples/hepsim/pythia6_zpole_tautau.py # run over HepSim Monte Carlo data
The last example from the HepSim web page prints the pT distribution of e, mu, taus. You can also run these examples as “fpad_edit file.py” or using the Jas-like environment (i.e. jaspp program).
You can find many examples from HepSim validation page (ProMC) and LCIO validation page.
Here is an example of how to compare data points with a histogram using DMelt classes. Run this script as “fpad file.py”, or open it in the Jas4pp editor and run this script.
Click to show the Python code
Click to show the Python code
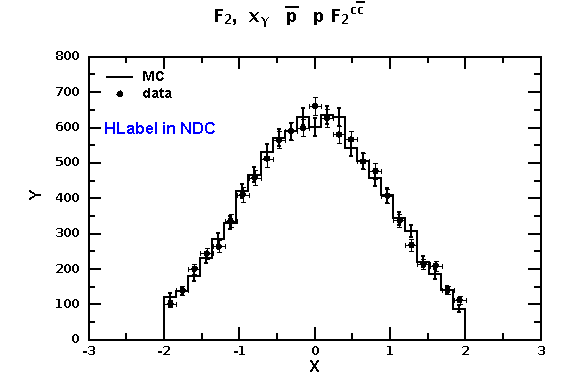
from java.awt import * from java.util import Random from jhplot import * c1 = HPlot() # c1.doc() # view documentation c1.setGTitle("F_{2}, x_{γ} #bar{p}p F_{2}^{c#bar{c}}") c1.visible(1) c1.setAutoRange() c1.setMarginLeft(85) # make space for Y label c1.setNameX("X") c1.setNameY("Y") h1 = H1D("MC",25, -2, 2.0) h1.setPenWidthErr(2) # line width h2 = H1D("data",25, -2, 2.0) r=Random() for i in range(10000): h1.fill(r.nextGaussian()) h2.fill(r.nextGaussian()) p1=P1D(h2) # convert to X-Y array c1.draw(h1) c1.draw(p1) # c1.drawStatBox(h1) # set HLabel in the normilised coordinate system lab=HLabel("HLabel in NDC", 0.18, 0.7, "NDC") lab.setColor(Color.blue) c1.add(lab) c1.update() c1.export("image.eps")
Run this example in Jas4pp and look at this image:
Supported file formats
Jas4pp supports the following file formats:
- ROOT6 (read only). Also ROOT versions 3,4,5 are supported for limited number of ROOT objects. XROOTD is included
- LCIO (read/write)
- ProMC (read/write)
- ProOI (read)
- HiPO file format (read/write)
- HepRep (read)
- AIDA XML-format, including compression (read/write)
- STDHEP
- All I/O file formats supported by Python 2.7 (or pure-Python external modules)
- All I/O file-format methods from Java
These file formats are natively supported in Jython/Python, Groovy and Java codes.
Reading ROOT files
Jas4pp natively reads commonly used objects and data structures from ROOT files (versions 3, 4, 5 and 6). ROOT files can be opened using the Jas4pp menu [File]-[Open data source]-[Root file] (*.root).
In addition, one can work with ROOT files using Jython/Python and Groovy scripts. One can find some examples in the directory “examples/root”. Here is a Jython/Python example showing how to read a simple ROOT tree:
Click here to show a Python example
Click here to show a Python example
from hep.io.root.interfaces import TTree from hep.io.root import RootFileReader reader = RootFileReader("ntuple_tree.root") tree = reader.get("tree"); maxevents=tree.getEntries() leaves = tree.getLeaves() nrleaves=leaves.size() print "Nr of events=",maxevents print "Nr of leaves=",nrleaves print "Leaves:" for l in xrange( nrleaves ): print "Leaf=",(leaves.get(l)).getName() print "Run over events" f0=leaves.get(0); f1=leaves.get(1); f2=leaves.get(2); for i in xrange(tree.getEntries()): print f0.getValue(i), f1.getValue(i), f2.getValue(i)
The example directory also shows how to read histograms. Similar examples can be made using Java or Groovy scripting.
The supported ROOT interfaces are:
Click here for details of ROOT interfaces here
Click here for details of ROOT interfaces here
TArrayC TArrayD TArrayF TArrayI TArray TArrayL TAttAxis TAttFill TAttLine TAttMarker TAxis TBasket TBranchClones TBranchElement TBranch TBranchObject TClonesArray TCollection TDatime TDirectory TFile TGraph TH1D TH1F TH1 TH2D TH2F TH2 TKey TLeafB TLeafC TLeafD TLeafElement TLeafF TLeafI TLeaf TLeafL TLeafObject TLeafO TLeafS TList TMap TNamed TObjArray TObject TProfile TSeqCollection TStreamerBase TStreamerBasicPointer TStreamerBasicType TStreamerElement TStreamerInfo TStreamerLoop TStreamerObjectAny TStreamerObject TStreamerObjectPointer TStreamerString TString TTree
There are other interfaces for reading complex root trees as those from Delphes fast simulations.
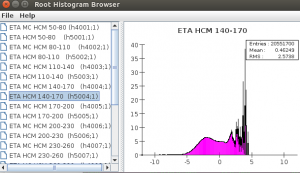
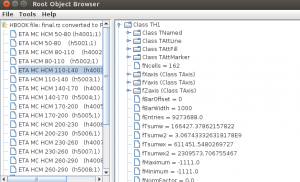
One can browser ROOT histograms (or other objects) using this:
import rootio rootio.HBrowser("histograms_root5.root") # browser and plot ROOT histograms rootio.Browser("histograms_root5.root") # browser for ROOT objects
You will see the browsers:
The last image shows the complete class structure of the ROOT file. Thses scripts can be put into Jython or Groovy files and executed as binary programs.
If the ROOT file has custom C++ classes, you need to create Java interface classes and compile them to the jar file and move this jar fil to “lib/user” directory of Jas4pp. To create interface classes for a ROOT file “test.root”, use:
import rootio rootio.Builder("test.root", None) # create interface classes in the current directory
This functionality is unchanged compared to FreeHep RootIO library.
Analysis of Delphes ROOT files
Jas4pp can be used for analysis of complex ROOT trees from fast simulations created by the Delphes framework. Many Delphes Monte Carlo files can be found in the HepSim repository. Download an example ROOT file with ttbar+jet events created with Madgraph5 from HepSim:
wget https://mc.hep.anl.gov/asc/hepsim/events/pp/13tev/mg5_ttbar_Njet/rfast102/mg5_ttbar_Njet_001.root
To study the structure of this ROOT file, run the browser:
import rootio rootio.Browser("mg5_ttbar_Njet_001.root")
The browser tells how the data are organized inside the ROOT file.
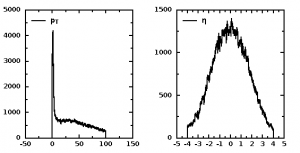
The process the tree “Delphes” from this file, one can write a small code in Python (writing in Groovy and Java is also trivial). Here is a simple code to show a histogram with particle ID:
Click here to show a Python code to read Delphes files
Click here to show a Python code to read Delphes files
from hep.io.root import RootFileReader xf="mg5_ttbar_Njet_001.root" # input file # examine file before doing anything # import rootio # rootio.Browser(xf) reader = RootFileReader(xf) tree = reader.get("Delphes") branches=tree.getBranches() print "Nr of events=",tree.getEntries() print "Nr of branches=",branches.size() for l in xrange(branches.size()): print "Branch=",l," name=",(branches.get(l)).getName() particles=tree.getBranch("Particle") # get Particle branch print "Type:", type(particles) branches=particles.getBranches(); print type(branches) for l in branches: print "particle ",type(l), " name=",l.getName() PID=particles.getBranchForName("PID") leaves=PID.getLeaves(); print PID.getName(), "has", leaves.size(); from jhplot import * h1 = H1D("particle ID",100, -10, 10) # fill a histogram pid=leaves.get(0) # get values with PID for all events for i in xrange(tree.getEntries()): h1.fill(pid.getValue(i)) c=HPlot("particle ID") c.visible() c.setAutoRange() c.draw(h1)
Note this code only works for integer values since we did not specify which value needs to be extracted.
If you need to extract double, float or boolean values, use the Delphes class and its “get” methods, getFloat, getDouble, getInt, and getBool. Here is an example that uses high-level classes to read transverse momentum (PT) and pseudorapidity (ETA) of Monte Carlo particles:
Click here to show a Python code to read Delphes files
Click here to show a Python code to read Delphes files
from hep.io.root import RootFileReader from rootio import * reader = RootFileReader("mg5_ttbar_Njet_001.root" ) tree = reader.get("Delphes") branches=tree.getBranches() print "Nr of events=",tree.getEntries() print "Nr of branches=",branches.size() print "Branches:" for l in xrange( branches.size() ): print "Branch=",l," name=",(branches.get(l)).getName() particles=tree.getBranch("Particle") # generator particles ptEvents=Delphes.getFloat(particles,"PT") # get PT array etaEvents=Delphes.getFloat(particles,"Eta") # get ETA array from jhplot import * h1 = H1D("PT of particles",100, 0, 100) h2 = H1D("Eta of particles",100, -4, 4) for i in xrange(tree.getEntries()): pt,eta=ptEvents[i],etaEvents[i] for j in xrange(len(pt)): h1.fill(pt[j]) h2.fill(eta[j]) c=HPlot("pT",600,300,2,1) c.visible() c.setAutoRange() c.draw(h1) c.cd(2,1) c.setAutoRange() c.draw(h2)
Histograms from Fortran and C++
Jas4pp can visualize histograms created by Fortran or C++ code (but without using ROOT). For this, use a light stand-alone library called CFBook (See CFBook). You can compile it using gcc (for C++ programs) or gfortran (for Fortran program). This library creates XML file with 1D and 2D histograms, that can be read by Jas4pp. Here is an example of reading 1D histogram from fortran.xml file:
Click to show the Python code
Click to show the Python code
from jhplot import * from jhplot.io import * hb = CFBook() hb.read("fortran.xml") print hb.listAll() print hb.getKeysH1D() # list keys h1=hb.getH1D(1) # use the key 1 to retrive H1D c1=HPlot("Test") c1.setGTitle("Histograms from a file"); c1.visible(1) c1.setAutoRange() c1.draw(h1)
Reading LCIO files
Jas4pp can be used to process LCIO files. It can read and write LCIO files. The examples are given in the directory “example”.
Click to show the Python code
Click to show the Python code
# Example of processing SLCIO files using Jas4pp # Setup Jas4pp using "source setup.sh". # Then create the directory "data" and fill it with SLCIO files using has-get command, # and run this script as "fpad example.py data" from org.lcsim.lcio import LCIOReader from hep.io.sio import SIOReader from hep.lcio.implementation.sio import SIOLCReader from hep.lcio.implementation.io import LCFactory from hep.lcio.event import * from hep.lcio.io import * from jhplot import * from hephysics.particle import LParticle import math import os,sys from java.lang import System; # get directory name from the argument filename = sys.argv[1] # make list of files.. import glob files = glob.glob(filename+"/*.slcio") factory = LCFactory.getInstance() nEvent=0 for f in files: print "Open file=",f reader = factory.createLCReader() reader.open(f) while(1): evt=reader.readNextEvent() if (evt == None): break nEvent=nEvent+1 # print " file event: ",evt.getEventNumber(), " run=",evt.getRunNumber() if (nEvent%100==0): print "# Event: ",nEvent col = evt.getCollection("MCParticle") colPF = evt.getCollection("PandoraPFOCollection"); colCl = evt.getCollection("ReconClusters"); colTr = evt.getCollection("Tracks"); colECB = evt.getCollection("EM_BARREL"); colHCB = evt.getCollection("HAD_BARREL"); nMc=col.getNumberOfElements(); nPF=colPF.getNumberOfElements(); nCl=colCl.getNumberOfElements(); nTr=colTr.getNumberOfElements(); nECB=colECB.getNumberOfElements(); nHCB=colHCB.getNumberOfElements(); # now you can access all data here # manage memory if too much data del col del colPF del colCl del colTr del colECB del colHCB del evt reader.close() # close the file del reader System.gc() # force memory cleanup, if needed
To process SLCIO files from a directory “data”, call this script “example.py as:
fpad example.py data
The script open each file, and access every container in the file. Note that we explicitly mange memory in case of very large files, forcing the garbage collector for each file. In many cases, this is not needed since JVM takes care of memory problems.
See LCIO validation page for more examples.
Reading LCIO files from Delphes
The Delphes program has somewhat different containers, but they alos can be processed by Jas4pp using Python code. You can find Delphes LCIO files in https://atlaswww.hep.anl.gov/hepsim/info.php?item=368 (created for Snowmass21). See rfast052 tag. To process such LCIO files, look at the example Python scripts in the directory “examples/delphes”.
Reading miniDST files
Jas4pp natively reads miniDST files created for the ILC studies. Here is the example showing how to make a Z-boson peak from a miniDST file. This file is described in this recent tutorial [https://indico.fnal.gov/event/45031/overview].
Here is the example:
wget https://atlaswww.hep.anl.gov/asc/jas4pp/data/rv01-16-p10_250.sv01-14-01-p00.mILD_o1_v05.E250-TDR_ws.I106479.Pe2e2h.eL.pR-00001-ILDminiDST.slcio
The run this code that reads this files and creates the Z-boson mass. The code is shown here:
show more details here
show more details here
from hep.lcio.implementation.io import LCFactory from hephysics.particle import LParticle from jhplot import HPlot,H1D from math import sqrt import glob files=["rv01-16-p10_250.sv01-14-01-p00.mILD_o1_v05.E250-TDR_ws.I106479.Pe2e2h.eL.pR-00001-ILDminiDST.slcio"] factory = LCFactory.getInstance() h1= H1D("Mass",50,0,200) # create a histogram h1.setFill(True) nEvent=0 for f in files: print "Open file=",f reader = factory.createLCReader() reader.open(f) while(1): evt=reader.readNextEvent() if (evt == None): break nEvent=nEvent+1 # print " file event: ",evt.getEventNumber(), " run=",evt.getRunNumber() if (nEvent%100==0): print "# Event: ",nEvent strVec = evt.getCollectionNames() if nEvent == 1: for col in strVec: print col col = evt.getCollection("IsolatedMuons") muons=[] for i in range( col.getNumberOfElements() ): track=col.getElementAt(i) mom=track.getMomentum() px,py,pz=mom[0],mom[1],mom[2] ee=sqrt(px*px+py*py+pz*pz) muons.append( LParticle("muon",px,py,pz,ee,0.105) ) col = evt.getCollection("Refined2Jets") jets=[] for i in range(col.getNumberOfElements()): jet=col.getElementAt(i) mom=jet.getMomentum() px,py,pz=mom[0],mom[1],mom[2] ee=sqrt(px*px+py*py+pz*pz) p=LParticle("jet",px,py,pz,ee,0) jets.append(p) if (len(muons)==2): # mass obj=muons[0] obj.add(muons[1]) mass=obj.calcMass() h1.fill(mass) #print(mass) del col,evt reader.close() # close the file del reader c1 = HPlot("=HepSim=",600,400) # plot histogram c1.setNameX("Mass") c1.setNameY("Events") c1.setSubTicNumber(0,5) c1.visible(True) c1.setAutoRange() c1.setRangeX(0,200) c1.setRangeY(0,10000) c1.setMarginLeft(90) c1.draw(h1)
Programming with HepSim
Jas4pp can be used to write Jython scripts to validate HepSim ProMC files. Please look at HepSim manual. All such java classes can be accessed using Java, JShell, Groovy and Jython programs.
Java API
One can learn about Jas4pp classes using Jas4pp API documentation. These classes can be used in Java, JShell, Groovy and Jython analysis programs.
Plots and histograms
Jas4pp uses histogram packages supported by both DMelt (community edition), JAIDA (FreeHep) or GROOT (JLab). DMelt provides programming API similar to PyROOT and with classes named conveniently to reduce code verbosity.
Here are a few most common classes:
- HPlot - canvas to show X-Y data and histograms in 2D
- HPlot3D - canvas to show X-Y-Z data and histograms in 3D
- H1D - 1D histogram
- H2D - 2D histogram
- P1D - X-Y container with support of 2-level errors
- j4np-physics - Physics vectors
The main canvas to show histograms H1D and data points P1D is HPlot. To process scripts in a background without a pop-up HPlot, use method “visible(False)”, and set sys.exit(0) at the end of the scripts. You can also use the JAIDA to make histograms (Histogram1D or Histogram2D). In addition, data are saved in the form of XML (with the extension ”.jdat“) files. Look the manual http://handwiki.org/wiki/DMelt:Start.
Lorentz particles and Jets
- PromcUtil convenient method to fill arrays with particles from ProMC files
- LParticle a HEP particle with the Lorentz transformations
- FastParticle a HEP particles with precomputed Et2,Eta,Phi for jet algorithms.
- Physics vectors typical HEP physics vectors the Lorentz transformations
- JetN2 recommended kt-type jet clustering algorithms (kT, anti-kT, CA) implemented in Java using N^2 approach, similar to the FastJet algorithm. Recommended for hadron collisions
- SCJet traditional kt-type jet clustering algorithms (kT, anti-kT) for pp implemented in Java using N^3 approach (slow)
- Jets and event shapes traditional jet algorithms and event shapes for e+e- from FreeHep (Geneva, Jade, Durham jets)
Java implementation of the longitudinally invariant kT and anti-kT clustering algorithms uses the E-scheme to combine particles (p1+p2) and Eta-Phi space (not Rapidity-Phi). Also, Cambridge/Aachen jets are supported. HepJet on github shows benchmarks of the Java implementation of the anti-KT with the original FastJet package.
You can build the standard kt-jets using PromcUtil, followed by the KTjet class.
Programming with GROOT
GROOT is a light-weight package for Java visualization created at JLAB. The full API of GROOT can be found in GROOT API page. There is also GROOT Wiki describing some features of GROOT.
Please look at the examples inside the directory “examples/groot”.
Visualizing events with Jas+Wired
You can run Jas+Wired to visualize the simulated events. The Wired program is included in Jas4pp, so you simply run it as:
./jaspp
This will start a Jas3-like environment with all needed plugins. Then copy the detector geometry file from the detector repository.
wget http://atlaswww.hep.anl.gov/hepsim/soft/detectors/sidloi3.tgz -O - | tar -xz;
This will create a directory “sidloi3”. This detector corresponds to “rfull001” tag used for the reconstruction of pythia6_zpole_ee (Z→e+e-).
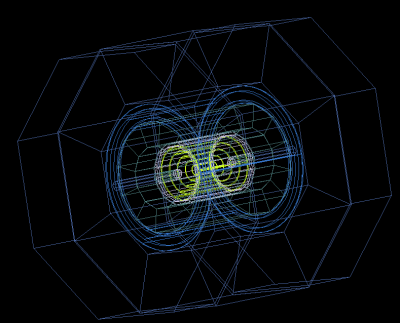
Now we can visualize the detector as [File]-[Open data source]-[HepRep] XML and select the file “sidloi3.heprep” from sidloi3.tgz: This is how to do this using the command line:
./jaspp sidloi3.heprep
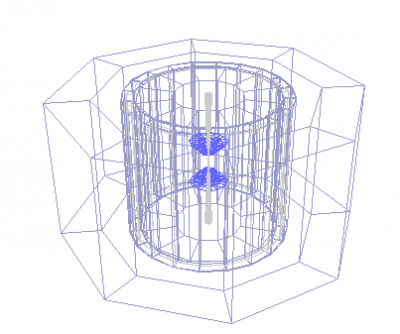
You will see the detector layout:
Now, we will read the event: Open any *.slcio file you copied from HepSim as [File]-[Open data source]-[LCIO] file. Then click a small button [Go] (top menu bar). It will process events. Then select again [File]-[New]-[Wired 4 view]. You will get an image in the Wired4 display. Now press [Go] again to look at next event.
If you want to see how data records are organized inside the slcio file, do this [File]-[New]-[LCSim Event browser]
Here is an example of how to visualize LCIO files from the HepSim repository
wget http://mc.hep.anl.gov/asc/hepsim/events/misc/pgun/pgun_eta35_muon//rfull009/pgun_muon1024gev_001_pandora.slcio ./jaspp pgun_muon1024gev_001_pandora.slcio
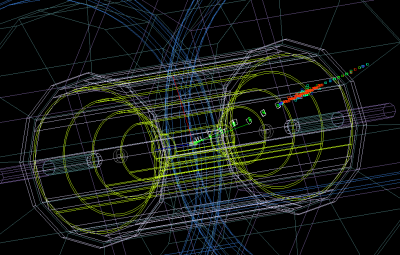
Click “Go” in the menu. Now you can open the event browser or Wired event display. Look at event record as “File→New→Event Browser”. Similarly, visualize the event as ” File→New→Wired4 View“. It shows an black window on the left. Then go to the next event, click “Go next” in the menu. This image illustrates a single electron event:
Similarly, you can view any events form the HepSim directory.
Generally, when you open a SLCIO file from the HepSim, you do not need to load the geometry file. The geometry file will be downloaded automatically from HepSim and will be put to ”.lcsim/cache/“ inside your home directory. Then go to the next event, click “Go next” in the menu. This image illustrates a single electron event:
Similarly, you can view any events form the HepSim directory.
Generally, when you open a SLCIO file from the HepSim, you do not need to load the geometry file. The geometry file will be downloaded automatically from HepSim and will be put to
.lcsim/cache
(inside your home directory).
Visualization of complex 100 TeV events are shown in the HepSim manual.
Visualizing geometry using ROOT
(contribution from N.Nikiforou):
You can also visualize the detector using ROOT. Here are a few steps: Locate the file detector.gdml inside the file detector.zip (where “detector” shows the name of the detector).
show more details here
show more details here
This file is created using this command during the detector design stage
slic -g detector.lcdd -G detector.gdml
Once you have the GDML file, you can use ROOT to visualize it. You just need to make sure the ROOT installation has openGL/GDML support (CERN AFS installations have it for sure). At the ROOT prompt do:
TGeoManager::Import("detector.gdml"); gGeoManager->GetTopVolume()->Draw("ogl");
This should popup an OpenGL display with the detector which you can clip, pan, rotate etc. You will the image as shown here:
Detector and physics studies
HepSim wiki explains how to use Jas4pp for physics and performance studies of future colliders. For example, this page shows how to visualize complex 100 TeV events. Such complex events were featured by this general article. Jas4pp is currently used for FCC, CEPC and EIC studies.
People
- S.Chekanov (ANL, main developer)
- E.May (ANL, debugging, Wired4)
- D.Blyth (speedup rendering of events and ProIO support)
- N.Nikiforou
- G.Gavalian (JLab)
- Khushi Taori (ANL SULI 2021 student). ROOT6 support validation
Additional help
- http://jas.freehep.org/jas3/ - JAS3
- http://java.freehep.org/ - FreeHep
- https://datamelt.org - DataMelt
- https://atlaswww.hep.anl.gov/asc/promc/ ProMC file format
- http://lcio.desy.de/ LCIO file format
Source code
Jas4pp is licensed under GPL. The source code available on Github (under construction).
— Sergei Chekanov 2016/03/23 21:53