Table of Contents
=HepSim=
HepSim is a public repository with Monte Carlo simulated events for high-energy physics (HEP) experiments, current and future. The HepSim repository was started at ANL during the US long-term planning study of the American Physical Society’s Division of Particles and Fields (Snowmass 2013) with the goal to create references to truth-level MC records for current and future experiments.
The repository can be used to validate and reproduce theoretical predictions (cross sections, distributions from particle 4-momenta) with full sets of theoretical uncertainties (scale, PDF, etc). Currently, it contains events from leading-order (LO) parton shower models, next-to-leading order (NLO) and NLO with matched parton showers.
Monte Carlo events are generated using the BlueGene/Q supercomputer of the Argonne Leadership Computing Facility ( BG@HepLib library), ANL-HEP cluster, ATLAS Connect virtual cluster service, OSG (Open Science Grid) Connect and NERSC. The file storages for this project are provided by ATLAS Connect, NERSC science gateways and by ANL HEP.
The Monte Carlo files can be accessed from the HepSim repository link.
Physics and detector studies
Here are several links to extending this Wiki for particular detector-performance topics:
- FCC-hh detector studies - explains how to analyses data for FCC-hh detector studies
- SID detector studies - explains how to analyses data for the SiD detector (ILC)
- CEPC detector studies - shows some results with full simulations for CEPC
- EIC detector studies - shows some results with full simulations for EIC
- HCAL studies explains how to analyse ROOT data after fast detector simulations used for FCC studies
Currently these studies are based on Jas4pp data-analysis environment. Geometry files for full detector simulations can be found in the detector repository.
This manual describes how to work with truth-level samples posted on HepSim. HepSim Python/Java analysis page explains how to write code to read truth-level files using Python on the Java platform.
<fc #008000>We are working on a more extensive manual here</fc>.
Data formats
Truth-level data are stored in a platform-independent format called ProMC that allows very effective compression using a variable-byte encoding. This data format is supported by popular programming languages (C++, Java, Python) on major operating system (Windows, Mac, Linux, etc.).
Simulated data after fast and full detector simulations are kept in ROOT and LCIO formats.
HepSim software toolkit
In order to work with HepSim database, install the HepSim software toolkit. For Linux/Mac with the bash shell, use:
wget http://atlaswww.hep.anl.gov/hepsim/soft/hs-toolkit.tgz -O - | tar -xz source hs-toolkit/setup.sh
or using curl:
curl http://atlaswww.hep.anl.gov/hepsim/soft/hs-toolkit.tgz | tar -xz source hs-toolkit/setup.sh
This creates the directory “hs-toolkit” with HepSim commands. To run them, Java7/8 and above should be installed. One can also download hs-toolkit.tgz. Most commands are platform-independent and based on the hepsim.jar that can be used on Windows/Mac. The setup command adds all commands to the execution path.
You can view the available commands from this package by typing:
hs-help
Event record browser
Use the event-record browser to view separate events, cross sections and log files. On Linux/Mac, use:
hs-view http://mc.hep.anl.gov/asc/hepsim/events/pp/14tev/higgs/pythia8/pythia8_higgs_1.promc
Here we looked at one file of the Pythia8 (QCD) sample.
Of course, one can look at the local file as well:
hs-view pythia8_higgs_1.promc
after you downloaded it.
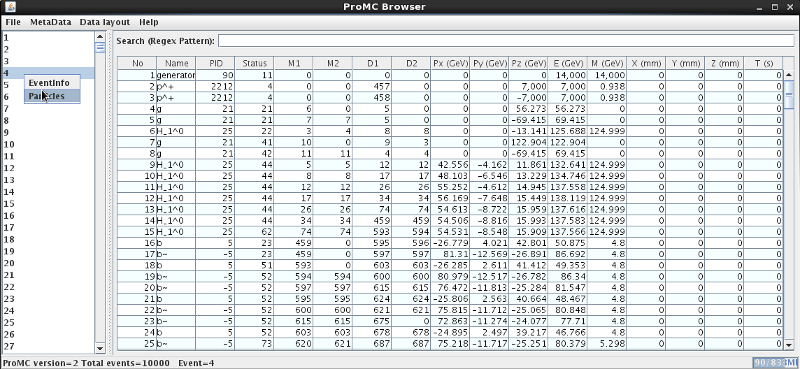
On Windows, download hepsim.jar and click the “hepsim.jar” file. Then open the ProMC file using the “File” menu. You will see a pop-up GUI browser which displays the MC record. You can search for a given particle name, view data layouts and log files using the [Menu]:
This works for full parton-shower simulations with detailed information on particles. Unlike the usual parton shower Monte Carlo, this browser has a detailed information on event weights, PDF uncertainties and scale uncertainties (in some cases). The browser can show 4-momenta of each event as well as the total cross sections (for NLO, you need to read all events to get an accurate cross section). Look at the ProMC file description.
File validation
One can check the consistency of the file and additional information as:
hs-info http://mc.hep.anl.gov/asc/hepsim/events/pp/14tev/higgs/pythia8/pythia8_higgs_1.promc
The last argument can be a file path on the local disk (faster than URL!). The output of the above command is:
File = http://mc.hep.anl.gov/asc/hepsim/events/pp/14tev/higgs/pythia8/pythia8_higgs_1.promc
ProMC version = 2
Last modified = 2013-06-05 16:32:18
Description = PYTHIA8;PhaseSpace:mHatMin = 20;PhaseSpace:pTHatMin = 20;ParticleDecays:limitTau0 = on;
ParticleDecays:tau0Max = 10;HiggsSM:all = on;
Events = 10000
Sigma (pb) = 2.72474E1 ± 1.92589E-1
Lumi (pb-1) = 3.67007E2
Varint units = E:100000 L:1000
Log file: = logfile.txt
The file was validated. Exit.
All entries are self-explanatory. Varint units - values used to multiply energy (momenta) to convert to variable-byte integers. The “E:100000” means that all px,py,pz,e,mass were multiplied by 100000, while all distances (x,y,z,t) were multiplied by 1000. See the ProMC archive format.
One can look at separate events using the above command after passing an integer argument that specifies the event to be looked at. This command prints the event number 100:
hs-info http://mc.hep.anl.gov/asc/hepsim/events/pp/14tev/higgs/pythia8/pythia8_higgs_1.promc 100
List available data files
Let us show how to find all files associated with a given Monte Carlo event sample. Go to HepSim database and find a sample by clicking on the “Info” column. Then find the files as:
hs-ls [name]
where [name] is the dataset name, or the URL of the Info page, or the URL of the location of all files. This command shows a table with file names and their sizes.
Here is an example illustrating how to list all files from the Higgs to ttbar Monte Carlo sample:
hs-ls tev100_higgs_ttbar_mg5
If you want to create a simple list without decoration to make a file to be read by other program, use
hs-ls tev100_higgs_ttbar_mg5 simple
The string “simple” removes the decorations. If you want a list with full URL and without decorations, use:
hs-ls tev100_higgs_ttbar_mg5 simple-url
Similarly, one can use the Info or Download URL path:
hs-ls http://atlaswww.hep.anl.gov/hepsim/info.php?item=2
or
hs-ls http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/higgs_ttbar_mg5/
The commands show this table:
Searching for a dataset
One can find a URL that corresponds to a dataset using a search word. The syntax is:
hs-find [search word]
For example, this commands list all datasets that have the keyword “higgs” in the name of the dataset, in the name of Monte Carlo models, or in the file description:
hs-find higgs
Note that there is no need to use any character such as “*” or “%” as for the usual regular expression. The match is done using any dataset that has the sub-string “higgs”. The same functionality is implemented in the “Search” menu of the HepSim interface.
If you are interested in a specific reconstruction tag, use “%” to separate the search string and the tag name. Example:
hs-find pythia%rfast001
It will search for Pythia samples after a fast detector simulation with the tag “001”. To search for a full detector simulation, replace “rfast” with “rfull”.
Downloading truth-level files
One can download all files for a given dataset as:
hs-get [name] [OUTPUT_DIR]
where [name] is either the name of the dataset, or the URL of Info page HepSim repository, or a direct URL pointing to the locations of ProMC files. This example downloads dataset “tev100_higgs_ttbar_mg5” to the directory “data”:
hs-get tev100_higgs_ttbar_mg5 data
Alternatively, this example downloads files using the URL of the Info page:
hs-get http://atlaswww.hep.anl.gov/hepsim/info.php?item=2 data
Or, if you know the download URL with the file locations, use this command:
hs-get http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/higgs_ttbar_mg5 data
All these examples will download all files from the “tev100_higgs_ttbar_mg5” event sample.
You can stop downloading using [Ctrt-D]. Next time you start it, it will continue the download. By default, we use 2 threads. One can increase the number of threads by adding an integer number to the end of this command. If a second integer is given, it will be used to set the maximum number of files to download. This example shows how to download 10 files using 3 threads:
hs-get http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/higgs_ttbar_mg5 higgs_ttbar_mg5 3 10
Instead of [Download URL], one can use the URL of the info page, or the name of the dataset. Here are 2 identical examples to download 5 files using single (1) thread and the ouput directory “data”:
Using the URL of the info page:
hs-get http://atlaswww.hep.anl.gov/hepsim/info.php?item=2 data 1 5
or, when using the dataset name given on the info page:
hs-get tev100_higgs_ttbar_mg5 data 1 5
You can also download files that have certain pattern in the names. If a directory contains files generated with different pt cuts, the names are usually have the substring “pt”, followed by the pT cuts. In this case, one can download such files as:
hs-get tev13_higgs_pythia8_ptbins data 2 5 pt100_
The last argument shows that all the downloaded files should have the string “pt100_” in their names (in this case, it tells that the file are generated with pT>100 GeV).
The general usage of the hs-get command requires 2, 3, 4 or 5 arguments:
[URL] [OUTPUT_DIR] [Nr of threads (optional)] [Nr of files (optional)] [pattern (optional)].
where [URL] is either info URL, [Download URL], or the dataset name.
It is not recommended to use more than 6 threads for downloads.
Downloading files after detector simulation
Some datasets contain reconstructed files after detector simulations. Reconstructed files are stored inside the directory “rfastNNN” (fast simulation) or “rfullNNN” (full simulation), where “NNN” is the version number. For example, tev100_ttbar_mg5 sample includes the link “rfast001” (Delphes fast simulation, version 001). To download the reconstructed events for the reconstruction tag “rfast001”, use this syntax:
hs-ls tev100_ttbar_mg5%rfast001 # list reco files with the tag "rfast001" hs-get tev100_ttbar_mg5%rfast001 data # download to the "data" directory
The symbol “%” separates the sample name (tev100_ttbar_mg5) from the reconstruction tag (rfast001). If you want to download 10 files in 3 threads, use this:
hs-get tev100_ttbar_mg5%rfast001 data 3 10
As before, one can also download the files using the URL:
hs-ls http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/ttbar_mg5/rfast001/ # list all files hs-get http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/ttbar_mg5/rfast001/ data
Plotting distributions
HepSim repository can be used to reconstruct any distribution or differential cross section. Many HepSim MC samples include *.py scripts to calculate differential cross sections. You can run them using downloaded ProMC files (in which case you pass the directory with *promc files as an argument). The second approach is to run *.py scripts on a local computer without downloading ProMC files beforehand. In this case, data will be streamed to computer's memory and processed using your algorithm.
You can create plots using a number of programming languages, Java, Python, C++, Ruby, Groovy etc. Plots can be done on any platform, without modifying your system. C++ analysis programs require ROOT and Linux.
Below we will discuss how to analyse HepSim data using Java, since this approach works on any platform (Linux, Mac, Windows) and does not require installation of any platform-specific program. As before, make sure that Java 7 and above is installed (check it as “java -version”).
Method I. Running in a batch mode without downloaded ProMC files
You can run validation scripts in a batch mode as:
wget http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/ttbar_mg5/macros/ttbar_mg5.py hs-run ttbar_mg5.py
Another approach is to use Jas4pp or DataMelt. Such programs give more flexibility and more libraries for analysis. In this example, we will run a Python script that downloads data from URL into the computer memory and runs it in a batch mode. First, you will need an analysis code from HepSim. Look at ttbar sample from Madgraph: ttbar_mg5. Find the URL location of the analysis script (“ttbar_mg5.py”) located at the bottom of this page. Then copy the URL link of the file *.py using the right mouse button (“Copy URL Location”). Let us make a calculation of differential ttbar cross section.
Here is how to process the analysis using Jas4pp:
wget http://atlaswww.hep.anl.gov/asc/jas4pp/download/current.php -O jas4pp.tgz tar -zvxf jas4pp.tgz cd jas4pp source ./setup.sh # takes 5 sec for first-time optimization wget http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/ttbar_mg5/macros/ttbar_mg5.py # get the HepSim script fpad ttbar_mg5.py # process it in a batch mode.
Similarly, you can use a more complex DataMelt:
wget -O dmelt.zip http://jwork.org/dmelt/download/current.php # get DataMelt wget http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/ttbar_mg5/macros/ttbar_mg5.py # analysis script unzip dmelt.zip ./dmelt/dmelt_batch.sh ttbar_mg5.py
(first time it will run slower since it needs to rescan jar files). In this example, data with the URL (given inside ttbar_mg5.py) will be downloaded to the computer memory. You can also pass URL with data as an argument and limit the calculation to 10000 events:
./dmelt/dmelt_batch.sh ttbar_mg5.py http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/ttbar_mg5/ 10000
If you want to see a pop-up canvas with the output histogram on your screen, change the line “c1.visible(False)” to “c1.visible()” and comment out “sys.exit(0)” at the very end of the “ttbar_mg5.py” macro. You can change the output format from “SVG” to “PDF”, “EPS” or “PNG”. Look at the Java API.
Method II. Running in a batch mode after downloading ProMC files
The above approach depends on network availability at the time when you do the analysis. It is more convenient to download data files first and run over the data. If all your ProMC files are in the directory “data”, run this example code from the DataMelt directory:
./dmelt/dmelt_batch.sh ttbar_mg5.py data
Here is a complete example: we download data to the directory “ttbar_mg5”, then we download the analysis script, and then we run this script over the local data using 10000 events:
wget http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/ttbar_mg5/macros/ttbar_mg5.py # get analysis scrip hs-get http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/ttbar_mg5 ttbar_mg5 ./dmelt/dmelt_batch.sh ttbar_mg5.py ttbar_mg5 10000
Similarly, you can use Jas4pp.
Method III. Running in a GUI mode
You can perform short validation analysis using an editor as:
hs-ide ttbar_mg5.py
Run this code by pressing “run”. This approach uses a light-weight editor built-in inside the hs-tool package.
For Jas4pp, you can start an editor, correct the script, and run it:
./jaspp ttbar_mg5.py # Open the script in the editor
Then, use the right mouse button and select “Run Python”. You will see the output.
For the DMelt IDE, you can also bring up a full-featured GUI editor as this:
./dmelt/dmelt.sh ttbar_mg5.py
It will open the Python script for editing. Next, run this script by clicking the image of green running man on the status bar (or press [F8]).
Method IV. Running in a GUI mode using URL dialog
If you use DMelt, you can run this code using a more conventional editor:
../dmelt/dmelt.sh
On Windows, click “dmelt.bat”. Then find a HepSim analysis script with the extension *.py. You can do this by clicking “Info” column in the HepSim database. For example, look at the ttbar sample from Madgraph ttbar_mg5. Find the URL location of the analysis script with the extension *.py located in the bottom of this page. Then copy the URL link of the file *.py using the right mouse button (“Copy URL Location”). Next, in the DataMelt menu, go to [File]→[Read script from URL]. Copy the URL link of the *.py file to the pop-up DataMelt URL dialog and click “run” (or [F8] key). The program will start running with the output going to “Jython Shell”. When processing is done, you will see a pop-up window with the distribution.
If you have already analysis file, you can load it to the editor as:
./dmelt/dmelt.sh ttbar_mg5.py
and run it using “run” or [F8] key.
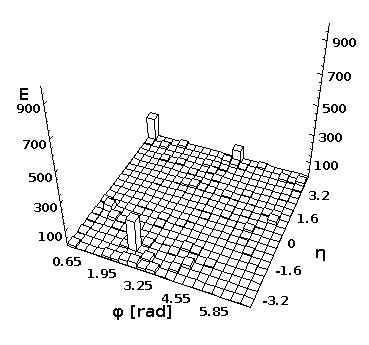
Event view in 3D
You can look at separate event and create a 2D lego plot for separate events using Python and Java. First, download any ProMC file, i.e.
wget http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/qcd_pythia8/pythia100qcd_001.promc
Then, execute this script in DatMelt which fills 2D histogram with final-state particles:
The execution of this script will bring-up a window with the lego plot.
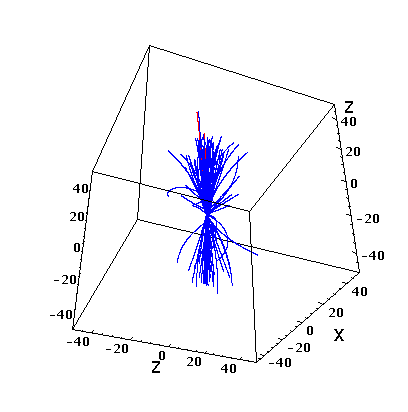
You can also look at a simple “tracking” for a given event:
The code written in Python is attached:
Reading NLO predictions
NLO records are different from showered MC. There is much less information available on particles, and events have weights. In many cases, PDF uncertainties are included. Here is an example how to read outputs from the MCFM program generated on BlueGene/Q at ANL.
hs-view http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/gamma_mcfm/gamma100tev_0000000.promc
or:
wget http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/gamma_mcfm/gamma100tev_0000000.promc hs-view gamma100tev_0000000.promc
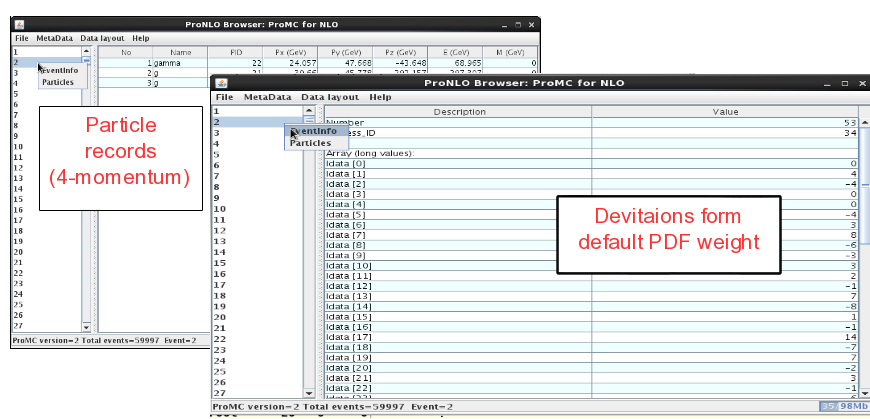
On the left panel, click on the event and then look at “Event info”. It shows the integer values (idata) that encode the PDF uncertainties, while the float array (“fdata”) shows other information. The first element in the float array is the weight of the event. The particle information is shown as usual (but without mother ID etc.).
The scripts that reconstruct cross sections are attached to the HepSim event repository. Here is how to run a script to reconstruct a direct-photon cross section at NLO QCD using DatMelt:
You can also open a script as:
wget http://mc.hep.anl.gov/asc/hepsim/events/pp/8tev/gamma_jetphox.py ./dmelt.sh gamma_jetphox.py
Alternatively, open this script in the DMelt editor and press run (or [F8]).
NLO event record includes 4-momenta of particles and event weights (double values). In addition, deviations form central weights are included as an array of integer values as:
You can calculate differential cross sections using online files using this example:
mkdir Higgs; cd Higgs; wget http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/higgsjet_gamgam_mcfm/macros/higgsjet_gamgam_mcfm.py wget -O dmelt.zip http://sourceforge.net/projects/dmelt/files/latest/download unzip dmelt.zip ./dmelt/dmelt_batch.sh higgsjet_gamgam_mcfm.py http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/higgsjet_gamgam_mcfm/
This example runs “higgsjet_gamgam_mcfm.py” code using online files and creates a Higgs differential cross section with PDF uncertainties. We use DMelt to do the calculations (after updating one jar file). You can also use ROOT/C++ to do the same.
Using C++/ROOT
To read downloaded ProMC files using C++/ROOT, you need to install the ProMC and ROOT package. If not sure, check these environmental variables:
echo $ROOTSYS echo $PROMC
They should point to the installation paths. If you use CERN's lxplus or AFS, simply one can setup PROMC as:
source /afs/cern.ch/atlas/offline/external/promc/setup.sh
which is built for x86_64-slc6-gcc48-opt.
Then, look at examples:
$PROMC/examples/reader_mc/ - shows how read a ProMC file from a typical Monte Carlo generator $PROMC/examples/reader_nlo/ - shows how read a ProMC file with NLO calculations (i.e. MCFM) $PROMC/examples/promc2root/ - shows how to read PROMC files and create ROOT Tree.
The same example directory shows how to write ProMC writes and convert to other formats.
You can generate an analysis code in C++, Java and CPython from a ProMC file with unknown data layout. Here is an example for a NLO file:
wget http://mc.hep.anl.gov/asc/hepsim/events/pp/8tev/gamma_jetphox/ggd_mu1_45_2000_run0_atlas50.promc promc_proto ggd_mu1_45_2000_run0_atlas50.promc promc_code make
This creates directories with the C++/CPython/Java analysis codes. For a longer description, read the ProMC manual.
For C++/ROOT, you can use this C++/ROOT example package. Untar it and compile using “make”. This will produce “promc2root” executable. It reads all ProMC files in a given directory and fills ROOT histograms with cross sections.
Use this Doxygen description to work with C++:
- annotated classes - for complete MC event records
- annotated classes - for NLO event records
Supported languages
- C++/ROOT
- CPython (Python implemented in C++) or PyROOT
- Java
- Jython (Python on the Java platform)
- Groovy (scripting language)
- BeanShell (scripting language)
- JRuby (Ruby on the Java platform)
Some variations of the Java languages are also possible (such as Scala). For all Java-derived languages, see the DatMelt project. The Android platform has a limited support (but the files should be readable on Android).
Although all example scripts are based on the Python language, the actual implementation of the validation online scripts is done in Jython (Java). You can program in Java as well. C++/CPython will be discussed later.
Using Jython
Jython and Java are chosen to write validation scripts and to run such scripts using web pages. Please look at HepSim Analysis Primer manual.
Using C++
Please refer ProMC web page on how to read/write ProMC files.
Also, there is a simple example showing how to read Monte Carlo files from HepSim in a loop, build anti-KT jets using FastJet, and fill ROOT histograms. Download hepsim-cpp package and compile it:
wget http://atlaswww.hep.anl.gov/asc/hepsim/soft/hepsim-cpp.tgz -O - | tar -xz; cd hepsim-cpp/; make
Read “README” inside this directory. Do not forget to populate the directory “data” with MC files before running this example.
Using CPython
Use the example above to generate CPython code to read data. This is documentation for CPython approach
- annotated classes - for complete MC event records
- annotated classes - for NLO event records
Read the description of ProMC format about how to generate CPython code.
Conversion to ROOT files
The ProMC files can be converted to ROOT files using “promc2root” converter located in the $PROMC/examples/promc2root directory. ROOT files will be about 30-50% larger (and their processing takes more CPU time). Please refer the ProMC manual.
cp -rf $PROMC/examples/promc2root . cd promc2root make ./promc2root [promc file] output.root
Fast detector simulation
On the fly reconstruction
HepSim can be used to create ROOT files after fast detector simulation, or one can analyse events after the Delphes fast simulation program on the fly. The latter approach allows to make changes to the detector geometry by the end-users and, at the the same time, perform an analysis. The output ROOT file only includes histograms defined by the user (but you can also add custom ROOT tree). To do this, use the FastHepSim package that includes Delphes-3.2.0 and ProMC 1.5, as well as an example analysis program. You can find a description of the current HCAL studies that use such an approach.
Follow these steps:
wget http://atlaswww.hep.anl.gov/asc/hepsim/soft/FastHepSim.tgz tar -zvxf FastHepSim.tgz cd FastHepSim/libraries/ ./install.sh
This installs “FastHepSim” which includes Delphes, ProMC and hs-tools to work with HepSim. Then setup the environment
cd .. # go to the root directory source setup.sh
Next, go to the analysis example:
cd analysis make
This compiles the analysis program (analysis.cc) that fills jetPT and muonPT histograms. Now we need to bring data from HepSim and put them somewhere. For this example, we will copy data to the “data” directory:
hs-get http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/higgs_ttbar_mg5 data 2 3
This copies 3 files in 2 threads and put them to the directory “data”. Now we are ready to run over these 3 files:
./Make_input data # creates inputdata.txt ./analysis delphes_card_FCC_basic_notau.tcl histo.root inputdata.txt
The first command creates a file “inputdata.txt” with input data. Then the program “analysis” reads the configuration file, data and generates “histo.root” with output histograms. This example uses “delphes_card_FCC_basic_notau.tcl” Delphes configuration file describing a basic FCC detector geometry. Note that we have removed tau tagging since we run over slimmed events with missing mothers of particles.
If you want to access other objects (photons, electrons, b-jets), use Delphes definitions of arrays inside the source code “analysis.cc”. You can put external files into the src/ directory where it will be found by Makefile.
If you still want to look at the event structure in the form of ROOT tree, run the usual Delphes command:
../libraries/Delphes/DelphesProMC delphes_card_FCC_basic_notau.tcl output.root data/mg5_Httbar_100tev_001.promc
where output.root will contain all reconstructed objects. In this case, add “TreeWriter” after “ScalarHT”. If the input file contains complete (non-slimmed) record, one can add “tau” reconstruction (“TauTagging” line).
Try also more sophisticated detector-geometry cards:
- examples/delphes_card_Snowmass_NoPileUp.tcl - similar to Snowmass 2013. It has CA (fat) jets and B-tagging
- examples/delphes_card_ATLASLIKE.tcl - similar to ATLAS detector (comes with Delphes)
Note that “TreeWriter” module should be enabled when creating ROOT files, and disabled when using analysis mode.
Creating Delphes files
Here we describe how to make fast detector simulation files using separate external libraries, without installing FastHepSim. Use the Delphes fast detector simulation program to process the MC events. Delphes can read ProMC files directly. First, make sure that ProMC and ROOT library is installed:
echo $PROMC $ROOTSYS
This should point to the installation paths of ProMC and ROOT.
Here are the steps to perform a fast detector simulation using ProMC files from the HepSim repository:
1) Download Delphes-3.2.0.tar.gz (or higher) and compile it as:
wget http://cp3.irmp.ucl.ac.be/downloads/Delphes-3.2.0.tar.gz tar -zvxf Delphes-3.2.0.tar.gz cd Delphes-3.2.0 ./configure make
This creates the converter “DelphesProMC” (among others), if “PROMC” environmental variable is detected.
2) For FCC studies, copy and modify the detector configuration file “delphes_card_FCC_basic.tcl”
cp cards/delphes_card_FCC_basic.tcl delphes_card_FCC_notau.tcl
Then remove the line “TauTagging”. Do the same when using the card “cards/delphes_card_ATLAS.tcl” (for ATLAS). We do not use the tau tagging module since it requires complete event records with all mother particles. Since ProMC files are often slimmed by removing some unstable low pT particles and showered partons, Delphes will fail on this line. If you need tau tagging, please use ProMC files with complete particle record.
3) Download Monte Carlo files from the 100 HepSim repository. For example, get a file with 5000 ttbar events generated for a 100 TeV collider:
wget http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/ttbar_mg5/mg5_ttbar_100tev_001.promc
and then create a ROOT file with reconstructed objects after a fast detector simulation (FCC detector):
./DelphesProMC delphes_card_FCC_notau.tcl mg5_ttbar_100tev_001.root mg5_ttbar_100tev_001.promc
The conversion typically takes 30 seconds.
Full detector simulation
Currently, the full detector simulation can be done using the SLIC software. You will need to convert ProMC files to LCIO files and use these files for the “sclic” Geant4-based program.
The physics performance studies are listed here. They are based on Jas4pp program.
How to convert ProMC to LCIO files is described in converting_to_lcio
Pileups mixing
One can mix events from a signal ProMC file with inelastic (minbias) events using a “pileup” mixing program:
hs-pileup pN signal.promc minbias.promc output.promc
Here “p” indicates that N events from minbias.promc will be mixed with every event from signal.promc using a Poisson distribution. If “p” is missing, then exactly N events from minbias.promc will be mixed with single event from signal.promc. Use large number of events in minbias.promc to minimise reuse of the same events from the minbias.promc. The barcode of particles inside output.promc indicates the event origin.
Here is an example to create events with pileup for the paper http://arxiv.org/abs/1504.08042 Phys. Rev. D 91, 114018 (2015): We mix h→ HH signal file with MinBias Pythia8 (A2 tune) sample using <mu>=200 (Poisson mean):
wget http://atlaswww.hep.anl.gov/asc/hepsim/soft/hs-toolkit.tgz -O - | tar -xz; source hs-toolkit/setup.sh wget http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/minbias_a2_pythia8_l3/tev100_pythia8_MinBias_l3__A2_001.promc wget http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/s0higgshiggs_alltau/tev100_s0higgshiggs_alltau_0001.promc hs-pileup p200 tev100_s0higgshiggs_alltau_0001.promc tev100_pythia8_MinBias_l3__A2_001.promc output.promc
Look at the “output.promc” with the browser to see how events are filled. Now you can use DELPHES to create fast simulation files using events with pileup.
Note: pileup mixer is implemented in Java, therefore it uses 64bit zip. For C++ programs, please use ProMCBook(“file.promc”, “r”, true) to open ProMC files (notice “true” as a new argument). For example, for the Delphes program, change the line
inputFile = new ProMCBook(argv[i], "r"); # 32bit zip
in readers/DelphesProMC.cpp to
inputFile = new ProMCBook(argv[i], "r", true); # 64bit zip
Extracting events
A file can be reduced in size by extracting N events as this:
hs-extract signal.promc N
where signal.promc is the original file, and N is the number of events to extract.
Comparing MC and data
HepSim maintains analysis scripts that can be used for comparing Monte Carlo simulations with data from Durham HepData database. For example, click the link with AAD 2013 (Search for new phenomena in photon+jet events collected in proton–proton collisions at sqrt(s) = 8 TeV with the ATLAS detector) paper
- Navigate to “DataMelt” and use right-mouse and select “Copy link location”
- Start DMelt if you did not yet, and select [File]-[Read script from URL]. Copy and paster the URL link from the HepData database
- Click “run”.
HepData maintain Jython scripts that use the same syntax as HepSim. You can start from a HepSim validation script, and before the “export” command, append the scripts with data from Durham HepData database. Or you can start from a HepData “SCaVis” script and add parts from HepSim validation script that reads data from HepSim. Note that SCaVis and DMelt are equivalent.
How to create ProMC files
There are several methods to create ProMC files from MC simulations. The most complete description is given here. Here are several examples:
- If you use Pythia8, look at the example “main46.cc” inside Pythia8 package.
- For other generators, such as Herwig++ and Madgraph, write events in either lhe files or hepmc files. Then use the converters “stdhep2promc” or “hepmc2promc” that are shipped with ProMC (inside $PROMC/examples).
If are working on a description of how to fill ProMC files from Jetphox or MCFM. At this moment, contact us to get help.
We have very limited support for other formats, since they are typically 1) large files (such as HEPMC); 2) cannot be used for streaming over the network; 3) are not suited in a multi-platform environments (i.e. Windows).
A note for ANL cluster
For ANL cluster, you do not need to install Delphes. Simply run the reconstruction as:
source /share/sl6/set_asc.sh $DELPHES/DelphesProMC delphes_card_FCC_notau.tcl mg5_ttbar_100tev_001.root mg5_ttbar_100tev_001.promc
The cards are located in $DELPHES/examples.
If you want to run over multiple ProMC files without manual download, use this command:
java -cp hepsim.jar hepsim.Exec DelphesProMC delphes.tcl output.root [URL] [Nfiles]
where [URL] is HepSim location of files and [Nfiles] is the number of files for processing. The output ROOT will be located inside the “hepsim_output” directory. Here is a small example:
java -cp hepsim.jar hepsim.Exec DelphesProMC delphes.tcl output.root http://mc.hep.anl.gov/asc/hepsim/events/pp/100tev/higgs_ttbar_mg5 5
which processes 5 files from Higgs to TTbar sample. Skip “5” at the end to process all files.
Single-particle gun
ProMC files with single particles can be created using the ProMC. Download the recent ProMC package and look at the directory:
wget http://atlaswww.hep.anl.gov/asc/promc/download/current.php -O ProMC.tgz tar -zvxf ProMC.tgz cd examples/particle_gun source setup.sh javac promc_gun.java java promc_gun pions.promc 100 211 1000 # E=0-100 GeV, PID=211, events=1000
This creates file “pions.promc” with 2 particles per event, with the maximum energy of 100 GeV (randomly distributed from 0 to 100). The particles are pions (pi+, pid=211). The total number of events 1000. The phi and theta distributions are flat After file creation, look at the events using the file browser. You can also modify the java code to change the single-particle events.
Converting to LCIO
ProMC files can be converted to LCIO files for full detector simulation. This is an example of such conversions:
wget http://atlaswww.hep.anl.gov/asc/promc/download/current.php -O ProMC.tgz tar -zvxf ProMC.tgz cd examples/promc2lcio source setup.sh javac promc2lcio.java java promc2lcio file.promc file.slcio
Look at other directories in “examples/”. You can convert ProMC files to many other formats (most converters require installation of the ProMC C++ package).
Record slimming
Particle records from the generators based on LO/NLO+parton showers calculations (PYTHIA, HERWIG, MADGRAPH) are often “slimmed” to reduce file sizes. In the case when records are slimmed, the following algorithm is used:
(status=1 && pT>0.3 GeV ) or # keep final states with pT>0.3 GeV (PID=5 || PID=6) or # keep b or top quark (PID>22 && PID<38) or # keep exotics and Higgs (PID>10 && PID<17) or # keep all leptons and neutrinos
where PID is absolute value of particle codes. Leptons ane neutrinos are also affected by the slimming pT cut. Note: for 100 TeV collisions, the pT cut is increased from 0.3 to 0.4 GeV. For NLO calculations with a few partons + PDF weights, the complete event records are stored.
In the case when the slimming is applied, file sizes are reduced by x2 - x3. In some situation, slimming can affect detector simulation. For example, you should turn of tau reconstruction in Delphes when slimming is used.
XML output format
Many scripts of HepSim create SVG images and a cross platform JDAT data format based on HBook. In order to convert JDAT files into CPython or ROOT object, read the data using xml.dom. This is a conversion to the CPython:
Snowmass13 samples
You can find more samples in the Snowmass MC web page.
How to cite
If you use HepSim event samples, Python/Jython analysis scripts and output XML files in your research, talks or publications, please cite this project as:
S.V. Chekanov. HepSim: a repository with predictions for high-energy physics experiments. Advances in High Energy Physics, vol. 2015, Article ID 136093, 7 pages, 2015. doi:10.1155/2015/136093 arXiv:1403.1886 and link. Use the bibtex entry:
@article{Chekanov:2014fga, author = "Chekanov, S.V.", title = "{HepSim: a repository with predictions for high-energy physics experiments}", year = "2015", eprint = "1403.1886", journal = "Advances in High Energy Physics", volume = "2015", pages = "136093", archivePrefix = "arXiv", primaryClass = "hep-ph", note = {Available as \url{http://atlaswww.hep.anl.gov/hepsim/}} }
Public results
Look at the list with pubic results based on HepSim using this Wiki
Contributions
Acknowledgement
The current work is supported UChicago Argonne, LLC, Operator of Argonne National Laboratory (``Argonne''). Argonne, a U.S. Department of Energy Office of Science laboratory, is operated under Contract No. DE-AC02-06CH11357. This research used resources of the Argonne Leadership Computing Facility at Argonne National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under contract DE-AC02-06CH11357.
Send comments to: — Sergei Chekanov (ANL) 2014/02/08 10:26